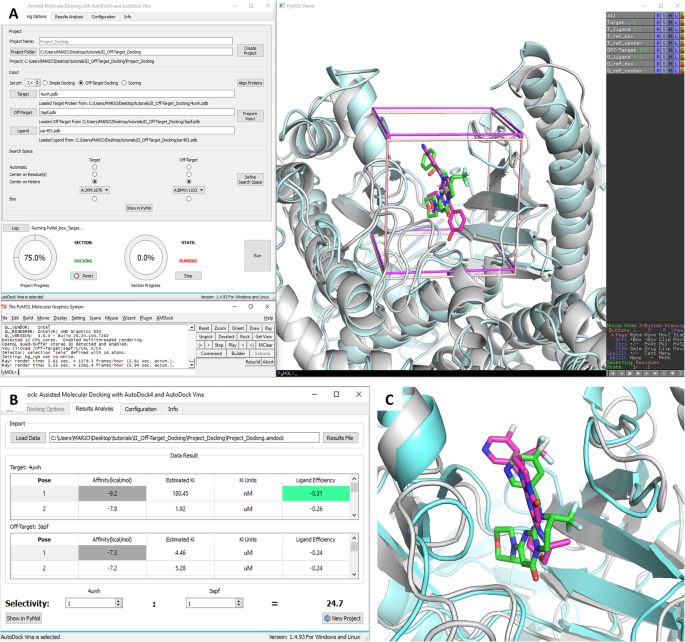
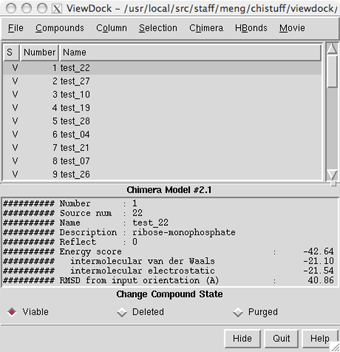
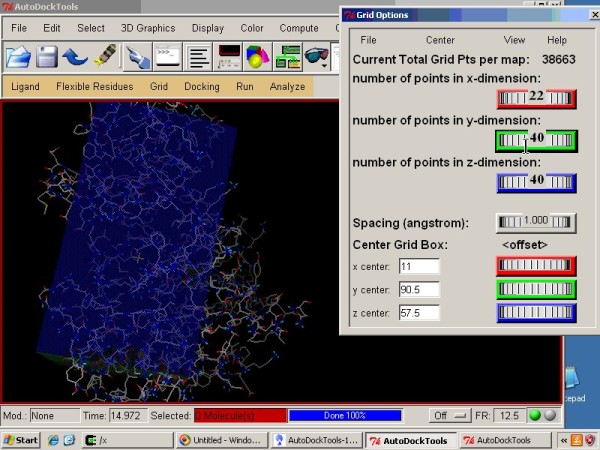
Molecular Docking Analysis | Autodock Results Analysis | Protein Ligand Int | Pymol | LigPlot Etc., - YouTube
Molecular Docking Analysis | Autodock Results Analysis | Protein Ligand Int | Pymol | LigPlot Etc., - YouTube

Molecular docking results employing AutoDock 4.2. Three dimensional... | Download Scientific Diagram

IJMS | Free Full-Text | DockingApp RF: A State-of-the-Art Novel Scoring Function for Molecular Docking in a User-Friendly Interface to AutoDock Vina

Evaluation of the binding performance of flavonoids to estrogen receptor alpha by Autodock, Autodock Vina and Surflex-Dock - ScienceDirect

Evaluation of the binding performance of flavonoids to estrogen receptor alpha by Autodock, Autodock Vina and Surflex-Dock - ScienceDirect

Improving AutoDock Vina Using Random Forest: The Growing Accuracy of Binding Affinity Prediction by the Effective Exploitation of Larger Data Sets - Li - 2015 - Molecular Informatics - Wiley Online Library

CrossDocker: a tool for performing cross-docking using Autodock Vina – topic of research paper in Biological sciences. Download scholarly article PDF and read for free on CyberLeninka open science hub.

Identifying Protein-Ligand Interactions with Colab | by JOSÉ MANUEL NÁPOLES DUARTE | Towards Data Science
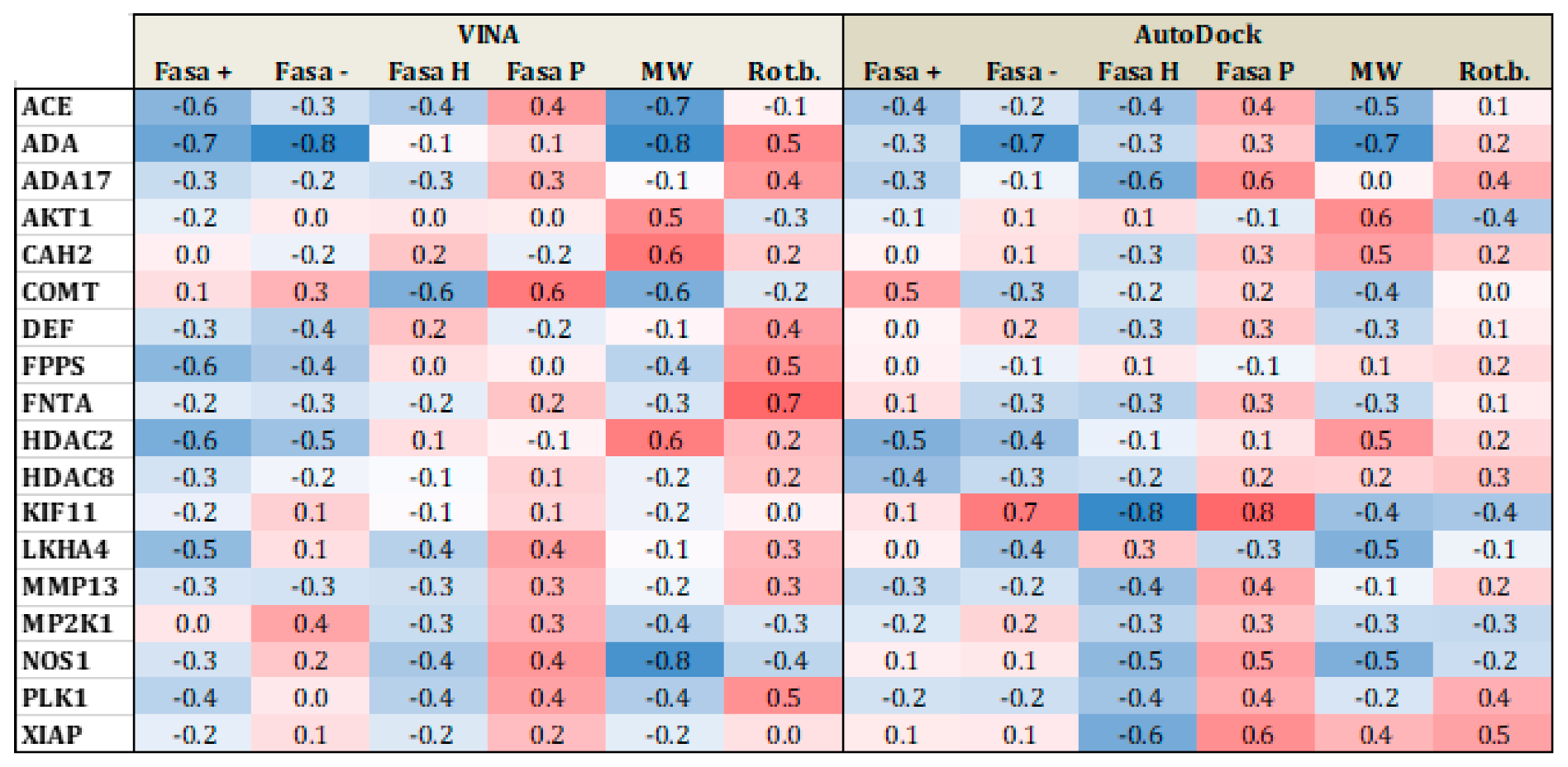
Applied Sciences | Free Full-Text | Comparing AutoDock and Vina in Ligand/Decoy Discrimination for Virtual Screening